Identification of Plasmid DNA
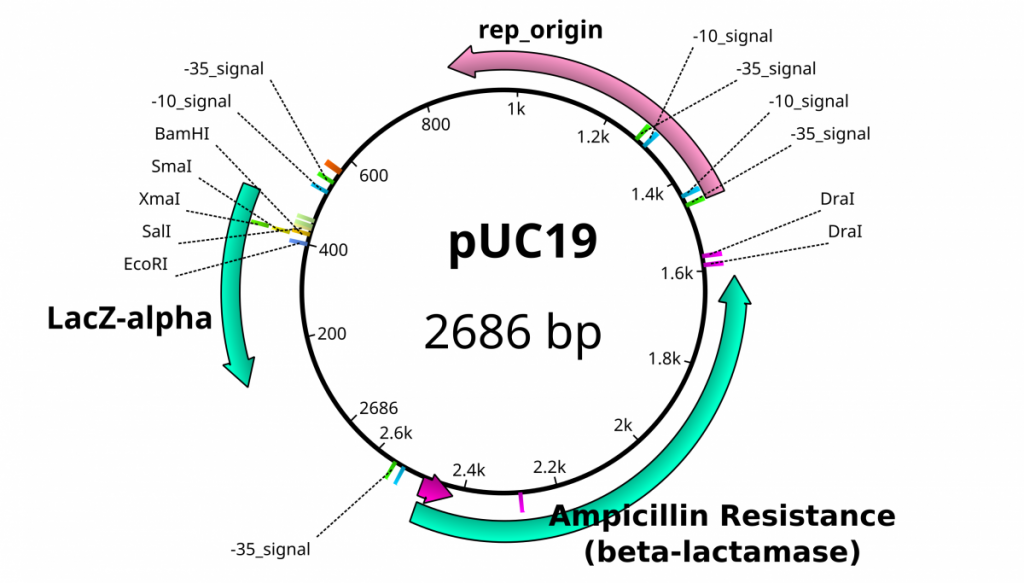
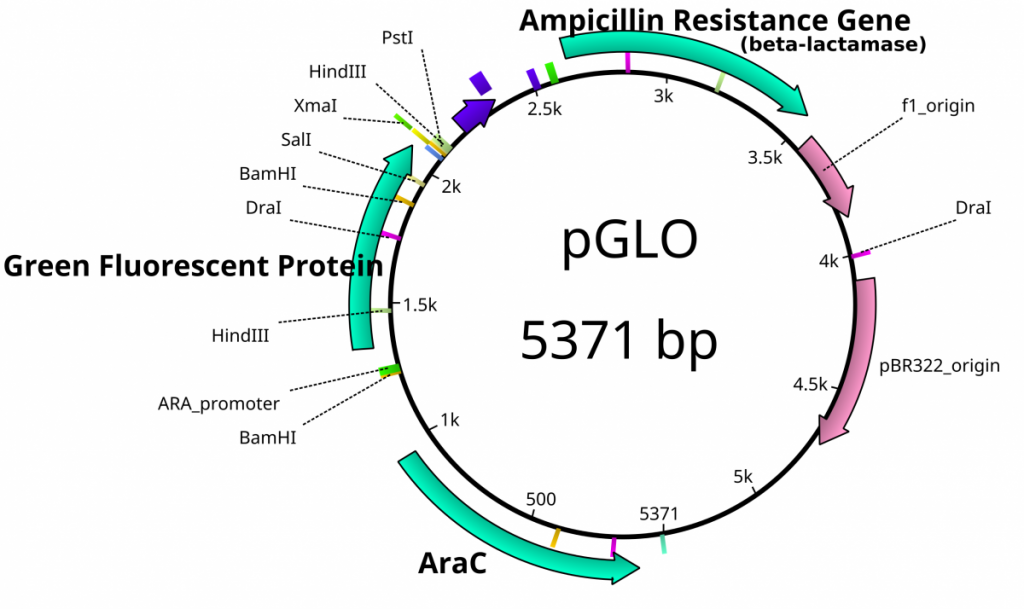
Once plasmids are isolated, they require identification. Plasmid vectors have known sequences and are mapped of their major features. Knowing the sequence of these pieces of DNA means knowing the locations of RE digestion sites. By using Res, digesting plasmids into known sizes aids in verification of plasmid identity without the need to have the entire plasmid re-sequenced. A common plasmid is called pUC18 or pUC19. The “p” stands for plasmid, the “UC” stand for University of California (where it was designed) and 18 or 19 refer to the difference in the MCS. This plasmid is 2,686 base pairs or ~2.7kb (kilobase) long with a single EcoRI site in the MCS. Another plasmid of interest in learning Molecular Biology is called pGlo. This plasmid has a jellyfish gene in the MCS that codes for a protein that will fluoresce green when expressed under UV light. pGlo is 5.4kb long and contains a single EcoRI site. Once digested, by an enzyme, these plasmids can be identified based on size separation on an agarose gel. Usually, it is best to identify by using 2 different REs. Digestion is important before size comparisons since circular DNA migrates through agarose differently than linear DNA. Additionally, circular DNA can sometimes be “super-coiled” and lead to very rapid migration despite the size.
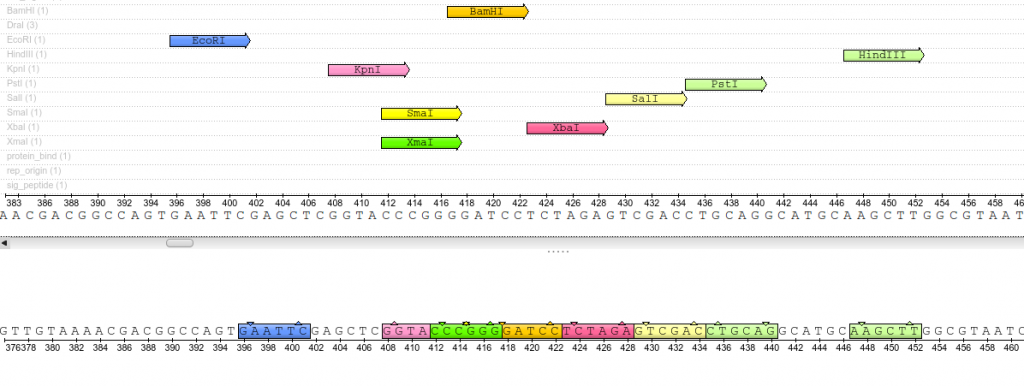
Close up view of the pUC19 Multiple Cloning Site (MCS). These restriction sites appear only once throughout the entirety of the plasmid sequence.
Exercise 2: Restriction Digestion Identification of Plasmids
- The class should prepare 2X 0.8% agarose gels by preparing 0.4g agarose in 50ml TBE buffer
- Melt agarose solution by microwaving for 1 minute
- Add 5μl Sybr Safe solution into 100ml gel solution
- Pour this solution into a casting tray inside the refrigerator
- insert a comb
- To a new tube add 2μl of plasmid DNA to 8μl of EcoRI fast digest mixture
- 1μl Fast Digest Buffer
- 1μl Fast Digest EcoRI enzyme
- 6μl H2O
- Incubate at 37°C for 10 minutes
- add 2μl of loading buffer to digestion mixture
- in a separate tube combine 3μl plasmid DNA with 2μl loading buffer and 7μl of H2O
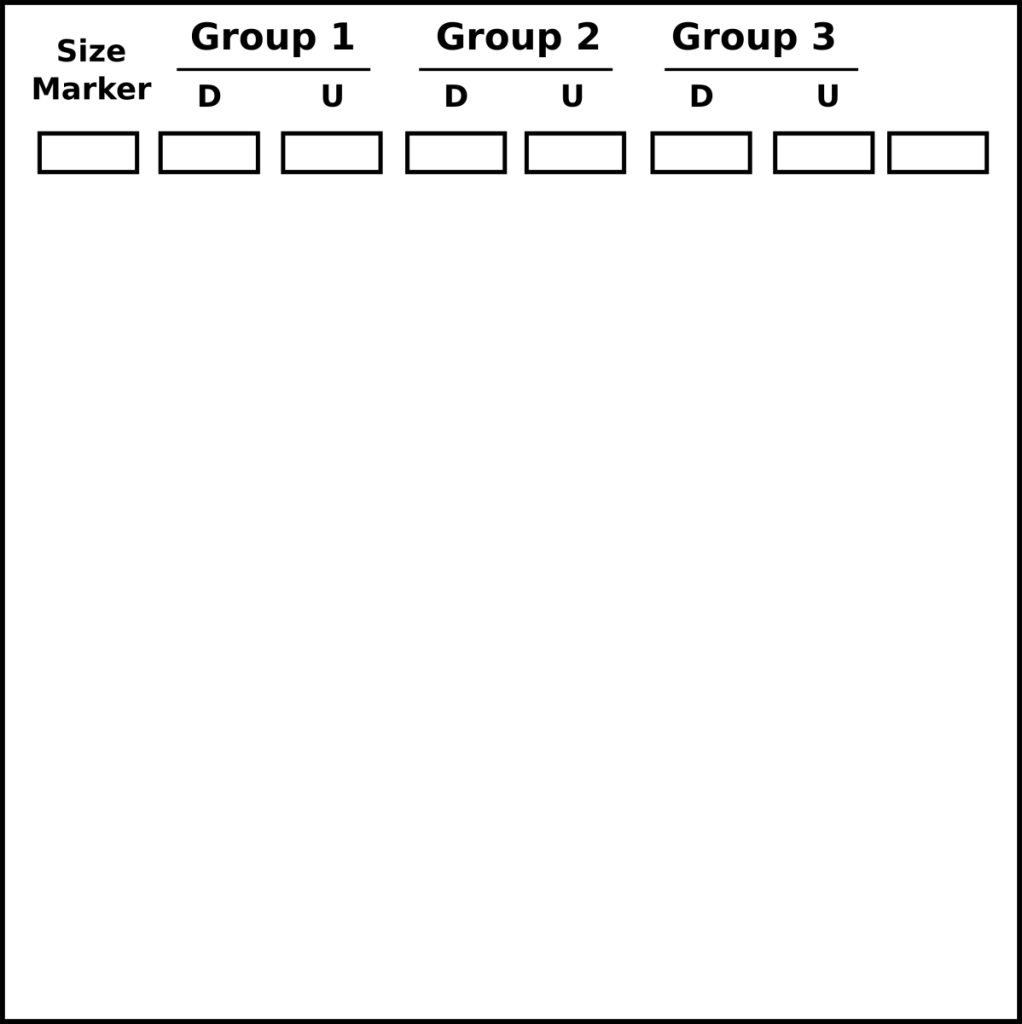
- Load gel with an appropriate size ladder in the first lane, in the next lanes load the Digested plasmid, then the undigested plasmid.
-
-
- 3 groups can load onto one gel
- U=undigested plasmid
- D=digested plasmid
-
-
- Run gel at 110V for 30 minutes and visualize on a UV transilluminator
- Document with your camera
Additional Resources
- For help on this problem, please try out the In silico digestion activity.